Overview
Since COVID-19 epidemic is still expanding around the world and posing serious threat to human life and health, we develop such a 2019nCoVAS webservice that not only offers online epidemic transmission prediction and LAUPs (Lineage-associated Underrepresented Permutations) analysis service to investigate the spreading trend and genome sequence characteristics, but also provides psychological stress assessment based on the emotion dictionary we build for 2019-nCoV.
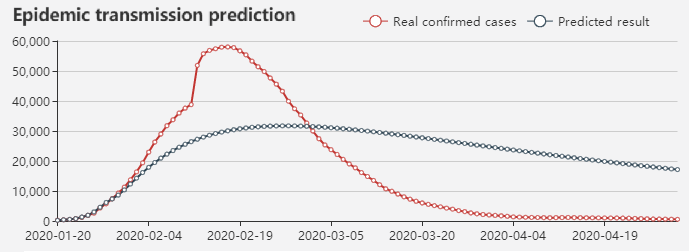
Epidemic transmission prediction
To predict the epdemic transmission and basic reproduction number(R0) for 2019-nCoV, we
offer three epidemic transmission predictive models: SIR, SEIR and SEIRQ. Especially,
the SEIRQ model considers quarantined case factor, which can be used to predict epidemic
situation under the condition of suspected cases quarantine. View details
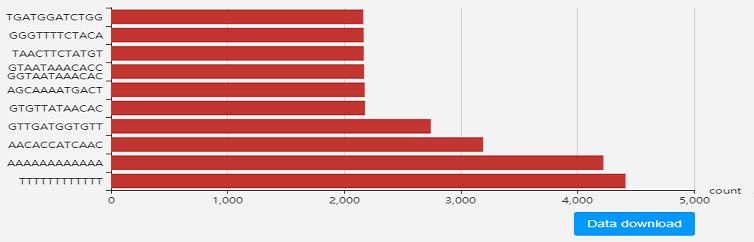
Genome sequence analysis
In order to analyze the LAUPs sequence characteris-tics for 2019-nCoV genome, we use JBLA
(Jellyfish-based LAUPs analysis application) to compute the underrepresented sequence
and common LAUPs. We also introduced the MOTIF discovery method to find the most
frequent arrangement pattern in the common LAUPs and provide data download service to
help users for further sequence and phylogenetic analysis. View details
Psychological stress assesment
We developed two corresponding features for our web server. One is to build an emotional
dictionary for 2019-nCoV, which can highlight its high frequent vocabulary. The other is
to assess and visualize the dynamic changes for the public's positive and negative
emotions in response to 2019-nCoV at different time point. These features not only are
able to provide the retrospective assessment function for psychologists, but also can be
used as the references for national policies development. View details

Citation
Xiao M, Liu G, Xie J, Dai Z, Wei Z, Ren Z, Yu J, Zhang L *. 2019nCoVAS: Developing the Web Service for Epidemic Transmission Prediction, Genome Analysis, and Psychological Stress Assessment for 2019-nCoV. IEEE/ACM Transactions on Computational Biology and Bioinformatics, 2021, DOI:10.1109/TCBB.2021.3049617.
Funding Support
This work was supported by National Science and Tech-nology Major Project [2018ZX10201002] and Sichuan University Postdoctoral Research and Development Foundation [2020SCU12056].
Contact Us
Address: College of Computer Science, Sichuan University. No. 24 South Section 1, Yihuan Road, Chengdu Sichuan, China, 610065.
E-mail: xiaoming@scu.edu.cn zhangle06@scu.edu.cn