Introduction
Exploring the relationship between CpG islands (CGIs) and methylation has become a hot spot in bioinformatics research. Although previous studies have made significant progress in developing methodology for CGI prediction, we are yet to evaluate the dynamics and complexity of all CpG sites, their clusters or CGIs, and their methylation status under variable conditions and dictated by enzymatic systems. Therefore, we build up the position-defined prediction method, investigate the connections between the methylation and CGI to further understand the methylation related regulatory mechanisms.
Human position-defined CGI prediction
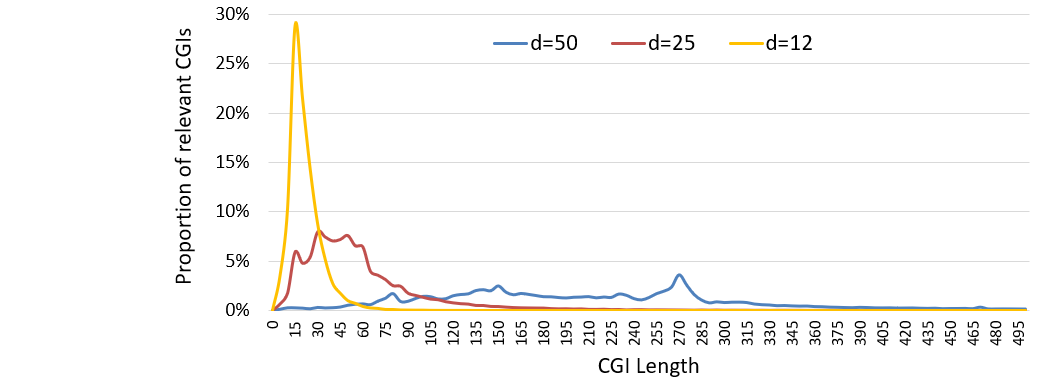
Quickly predict more Human short position-defined CGIs that may have unique location- or sequence- sensitive features and explore the differences in the genetic characteristics of CGIs predicted by different parameters.
Position-defined CGI prediction
CpG sites annotation analysis
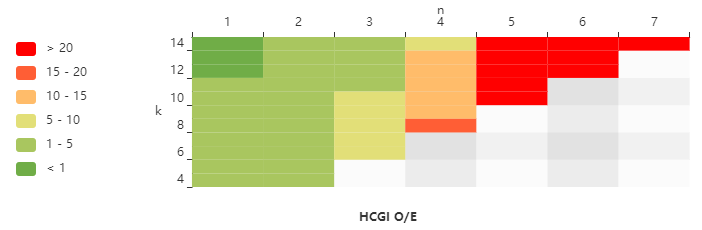
We introduce a novel human genome annotation and analysis protocol based on a combination of CGI density classification, gene expression breadth, and genome-wide methylation level annotation to investigate the connections between CGI, gene expression and methylation.
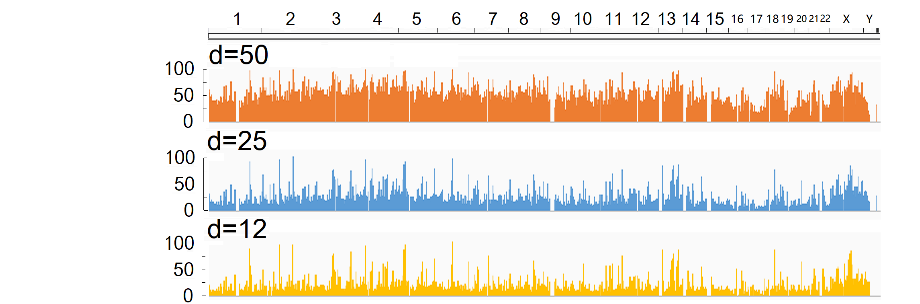
Human CpG sites Distribution analysis
CGI methylation analysis
Analyze the specificity of methylation level for CpG sites under different annotation categories.
Funding Support
This work was supported by grants from National Science and Technology Major Project (Grant No. 2018ZX10201002, China), China Postdoctoral Science Foundation (2020M673221, China), and Fundamental Research Funds for the Central Universities (2020SCU12056, China).
Contact Us
Address: College of Computer Science, Sichuan University. No. 24 South Section 1, Yihuan Road, Chengdu Sichuan, China, 610065.
E-mail: xiaoming@scu.edu.cn zhangle06@scu.edu.cn